Why we built it
Like much of the software world, the number of software solutions in life science has exploded in the last few years. Where the only option was once “good luck, build it yourself”, there are now a multitude of software options for life science R&D. This has created a totally new set of problems: software startups building similar tools, the software ecosystem becoming increasingly fragmented and non-interoperable, and biotech startups facing a glut of options and asking themselves “which software should we adopt?”* The biotech world is drowning in a sea of software products and tools, with few resources to help them figure out where to start or how to navigate options.
We felt it too – so we set out to map the landscape in hopes of finding out what software is available and how it can help. We had two specific goals:
- Understand what products exist and where there were gaps
- Create a starting-point resource for biotechs to find tools that might help them
The result is the Life Science Software Landscape (LSSL), a map of software for early-stage life science R&D. This map is the result of our work and feedback from the Bits in Bio community**, with contributions from Natalie Ma, Jonathan Karr, Ben Siranosian, Michael Makar, Vandon Duong, Gautam Chintapenta, Jason Sunardi, Ted Ling Hu, and Nicholas Larus-Stone.
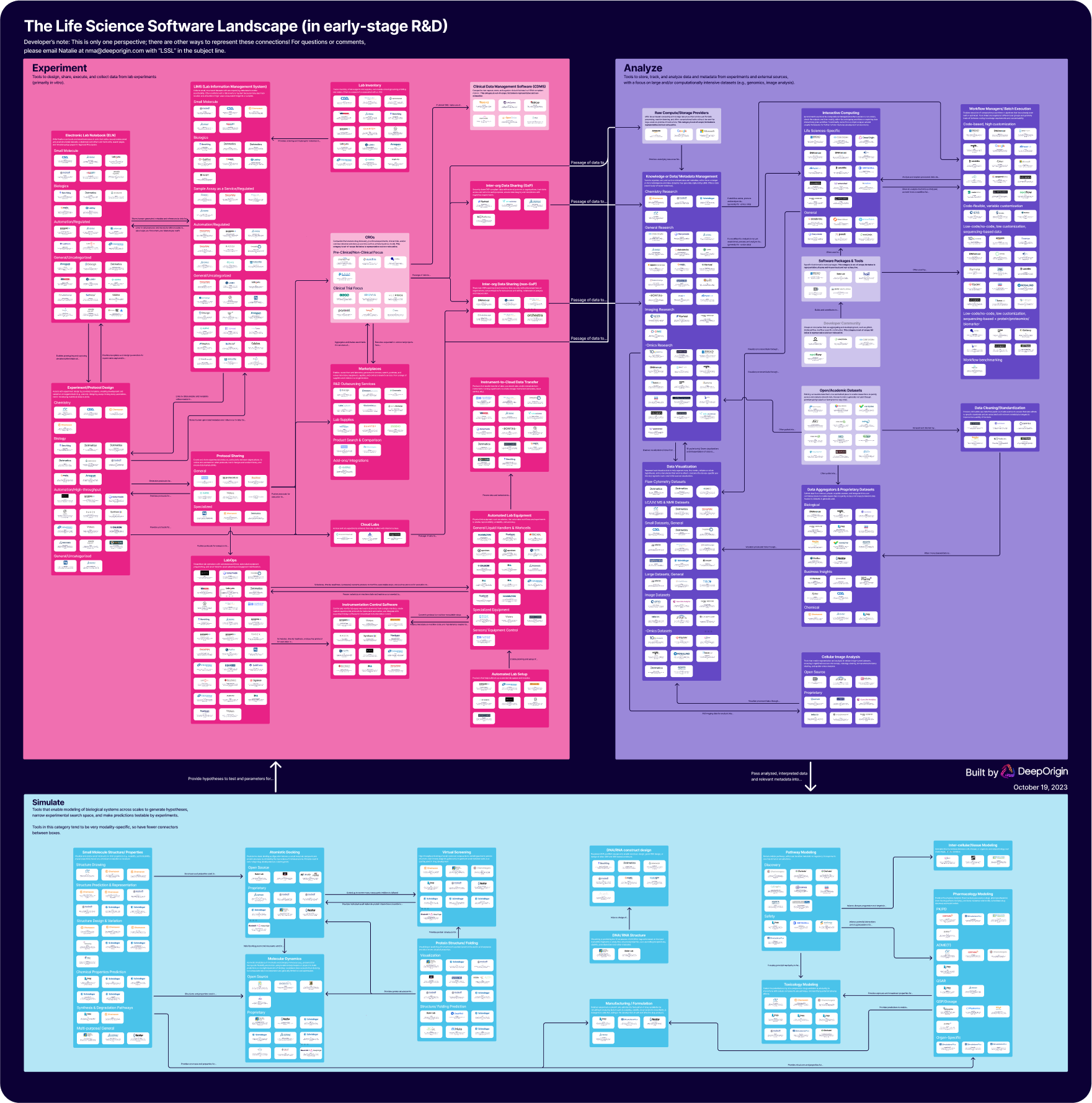
Lay of the Land(scape)
Ultimately, any software product needs to help each of us achieve our missions. Our mission at Deep Origin is to simulate living systems, from atoms to cells, to find causes of and treatments for diseases. For this to be possible, we (and the biotech community in general) needs an ecosystem of interoperable software tools across three areas: models of systems (i.e., simulations) to generate hypotheses, experiments to gather data, and analysis of data to test hypotheses - which then feeds back into models that generate new hypotheses and determine what experiment to run next. So we divided and classified software into three areas:
- Simulate (cyan): the modeling or simulation of biology, biological molecules, and biological processes or systems
- Experiment (fuchsia): the execution of experiments to collect data
- Analyze (purple): the analysis of data from experiments to test hypotheses
Areas we wanted to represent, but did not focus on in-depth are colored gray.
Within each of these areas, we defined categories of software based on goal or function. White boxes in these categories represent individual products that help a user achieve this goal. Where applicable, we further subcategorized products by focus (e.g., biologic versus small molecule modalities, sequencing versus imaging data).
Stepping back, it’s clear that many companies build products to collect and analyze data. Fewer companies connect modeling for hypothesis generation to conducting experiments to test them, or to analysis of data to update models of the world and generate novel hypotheses, or to connect all three verticals together. But completing this “simulate - experiment - analyze” loop is what will generate the most value in biotech, allowing us to more rapidly test hypotheses and re-use past data to inform the future. And the community needs solutions that connect these areas, not products that force people into islands of knowledge.
Where do we go from here?
Knowing what software is available and understanding what they do is just a first step. Despite the explosion of software products, gaps and challenges remain in technology’s service of life science R&D. An electronic lab notebook (ELN) and/or lab information management system (LIMS) alone cannot enable the next generation of breakthroughs in human health. A single bioinformatics platform or molecular dynamics tool won’t give us a full understanding of biology and the ability to predict experimental outcomes in silico. No one company will solve every problem. It will take a community, building both tools and mechanisms for them to connect and share information through APIs and protocols, to make this possible.
We would love your thoughts on this - you can contact us here.
Footnotes
*Our team has faced this problem at prior startups. We wondered what the options existed for running bioinformatic workflows and which would be best for our team. We struggled to figure out how to implement our ELN and LIMS: was Benchling really the best option, or was there a better option for our work? Product marketing rarely helped. And it was hard to find the time to dive deeply into the software options and evaluate them.
** We are working with the Bits in Bio community to create a community-maintained fork as a public resource for everyone building life science software.



.png)
.png)


.png)

.png)

.png)
.png)

.png)





.png)
.png)





.png)
.png)


.png)